The major mandate of "PAS" research group at the University of Jordan is to functionally apply holistic research approaches to dissect adaptation to abiotic stresses affecting major crop plants. Both molecular and physiological responses are studied and utilized to generate potential applicable resources and solutions to contribute to global sustainability and food security.
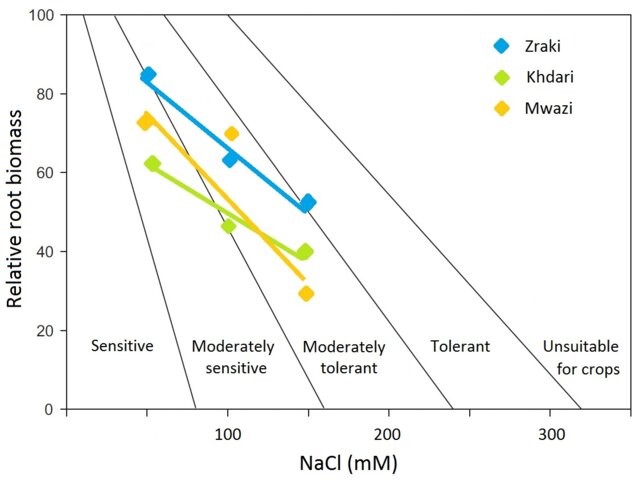
The research group identify major and novel differentially expression genes between top common fig cultivars from Jordan, e.g.
FcSORD and
FcDHN. Both molecular and physiological indicators endorse 'Zraki' cultivar as moderately tolerant against salinity stress.
![]()
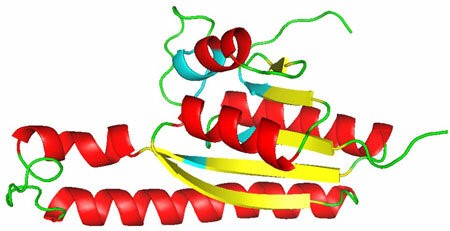
In another Mediterranean crop, our group discovered novel genes providing salinity tolerance in olive international cultivars, e.g. 'Nabali' from Jordan and 'Picual' from Spain. Among them,
OeUSP2 showed 6-8 fold increase in gene expression at 60 mM NaCl stress level. The predicated OeUSP2 three dimensional structure revealed unique ATP binding site utilizing residues from multiple helices and sheets.
![]()
In similar direction, the historical olive cultivar 'Mehras' represented by 1000-year old trees has been mitigating ages after ages of climate change and harsh environmental conditions. A collaborative national project was launched to decipher the genetic code of 'Mehras' applying cutting-edge next-generation sequencing technology. Millions of unique mutations were recorded and many were evident in the genetic coding regions, giving 'Mehras' the ability to survive ages of extreme environmental stresses while maintaining a distinctive olive oil quality. Both plastome and mitogenome showed that 'Mehras' is one of the oldest genetic olive genotypes in the Mediterranean region. This makes 'Mehras' an international indispensible genetic resource as a major genetic resource for breeding and improving olive cultivars that would have elevated tolerance capabilities compatible with climate change. The phylogenetic analysis showed that Mehras was genetically the closest to be a source of origin for the most important cultivated olives in the Mediterranean basin; namely 'Manzanilla' from Spain and 'Frantoio' from Italy. Therefore, 'Mehras' is a genuine ancestor variety, which has survived through the ages.


![]()
Another major research finding we achieved is related to memory genes and how they would help in mitigating abiotic stresses in different plan crops. In durum wheat, the regional main cultivar 'Hourani 27' was found to have the most robust epigenetic memory as a tool to withstand recurrent drought stress, which makes it a very resilient cultivar under dramatic climate conditions. Related work is being conducted in tomatoes.
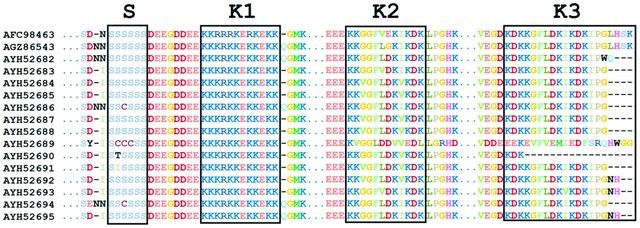
On the other hand, we are investigating one of the most halophytic plants, the Mediterranean saltbush (Atriplex halimus L.). Both the entire genome and transcriptome are being sequenced and processed. Several novel genes have been annotated and being investigated under salinity and drought stresses. One example applies to
AhDHN, with novel residues in multiple segments.
![]()
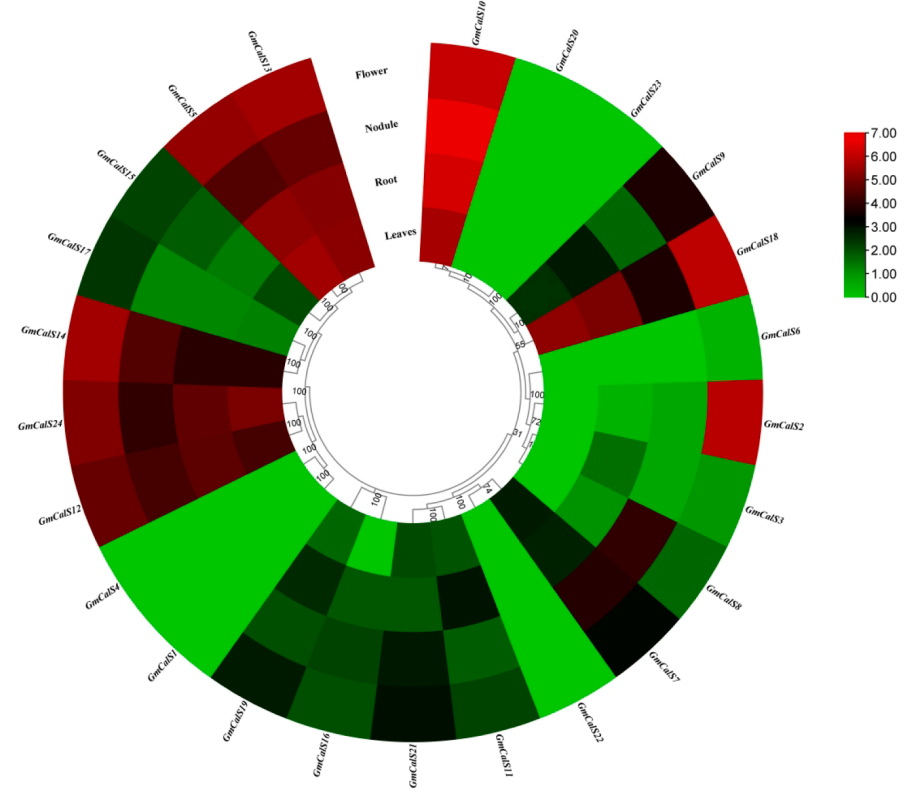
One major internationally important research theme is how crop plants develop under stresses. In this regard, several gene families were being investigated in soybean, a major global cash-crop.
![]()
Wild crop relative are major source of abiotic stress tolerance genes. To valorize such genes, genomic introgression is the most reasonable option to go with. In this regard, we have generated several introgression families between watermelon and desert bitter apple. After few generations, higher levels of TSS and lycopene content persevered and exceed parents.
References:
- Haddad, N., Migdadi, H., Brake, M., Ayoub, S., Obeidat, W., Abusini, Y., Aburumman, A., Al-Shagour, N., Al-Anasweh, E. and Sadder, M. (2021). Complete chloroplast genome sequence of historical olive (Olea europaea subsp.
europaea) cultivar Mehras, in Jordan. Mitochondrial DNA Part B, 6(1), 194-195.
- Musallam, A., Abu-Romman, S., & Sadder, M. T. (2023). Molecular Characterization of Dehydrin in Azraq Saltbush among Related Atriplex Species. BioTech, 12(2), 27.
- Sadder, M. T., Ateyyeh, A. F., Alswalmah, H., Zakri, A. M., Alsadon, A. A., & Al-Doss, A. A. (2021). Characterization of putative salinity-responsive biomarkers in olive (Olea europaea L.). Plant Genetic Resources, 19(2), 133-143.
- Sadder, M. T., Alshomali, I., Ateyyeh, A., & Musallam, A. (2021). Physiological and molecular responses for long term salinity stress in common fig (Ficus carica L.). Physiology and Molecular Biology of Plants, 27, 107-117.
- Sadder, M., Brake, M., Ayoub, S., Abusini, Y., Al-Amad, I., & Haddad, N. (2023). Complete mitochondrial genome sequence of historical olive (Olea europaea Linnaeus 1753 subsp.
europaea) cultivar Mehras in Jordan. Mitochondrial DNA Part B, 8(11), 1205-1208.
- Sadder, M. T., Musallam, A., Allouzi, M., & Duwayri, M. A. (2022). Dehydration Stress Memory Genes in Triticum turgidum L. ssp. durum (Desf.). BioTech, 11(3), 43.
- Zaynab, M., Sharif, Y., Xu, Z., Fiaz, S., Al-Yahyai, R., Yadikar, H. A., Al Kashgry, N.A.T., Qari, S.H., Sadder, M & Li, S. (2023). Genome-Wide Analysis and Expression Profiling of DUF668 Genes in Glycine max under Salt Stress. Plants, 12(16), 2923.
- Zaynab, M., Xu, Z. S., Yadikar, H. A., Hussain, A., Sharif, Y., Al-Yahyai, R. Sadder, M., Aloufi, A.s. & Li, S. (2023). Genome-Wide Analysis and Expression Profiling of CalS Genes in Glycine max Revealed their Role in Development and Salt Stress. Journal of King Saud University-Science, 103049.